Use of discovery proteomics as a biocompatibility screening tool: study of silver nanoparticle cytotoxicity
-
1
U.S. Food and Drug Administration, Center for Devices and Radiological Health, United States
Introduction: In vitro biocompatibility screening of devices is limited to assays for the evaluation of cytotoxicity, genotoxicity, blood interaction, and chemical characterization. These assays provide little information about the effects of the material on specific biological pathways and do not describe detailed biological effects at sub-toxic concentrations. The purpose of this work is to develop methodology and to demonstrate a proof-of-concept for the use of liquid chromatography (LC) mass spectrometry (MS) proteomics as a biocompatibility screening tool for new and emerging classes of biomaterials.
Silver (Ag), in both ionic and nanoparticle form, is known for its antimicrobial properties and has been widely used in various industrial and medical applications[1]. Silver nanoparticles (AgNPs) have emerged as an important class of nanomaterials for a wide range of medical devices, but there has been concern over possible Ag-induced toxicity[2]. Here we aim to quantitate the changes in protein amounts upon treating human cells with silver nanoparticles and to identify differentially regulated pathways using discovery proteomics.
Methods: Human monocyte cells (THP-1) were cultured using the stable isotope labeling by amino acids in cell culture (SILAC) technique[3] (Figure 1). Cells were activated to macrophage-like cells using phorbol 12-myristate 13-acetate (PMA) and treated with LC50 concentrations of AgNPs, silver nitrate [(AgNO3), ionic silver control], and gold nanoparticles [(AuNPs), nanoparticle control]. Processed peptides were subjected to LC-MS/MS on an Agilent 6540 QTOF system with a nanospray interface. The MS/MS data were searched against the Swissprot database using Agilent Spectrum Mill search engine for protein identification and differential expression quantitation. Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software.

Figure 1. Experimental workflow illustrating SILAC labeling and analysis methodologies.
Results and Discussion: We successfully identified and quantified a large number of proteins in cell extracts from each treatment group (398 to 739 per treatment with protein identification score >10 and relative standard deviation within acceptable limits). Of those identified, about 60 proteins exhibited an expression ratio of greater than 2, indicating a significant change compared to the untreated control. Preliminary pathway analysis has revealed that expression profiles among the samples is common to cellular growth and proliferation, cell death and survival, and protein synthesis, but unique expression profiles were also identified. For example, AgNP-treated cultures appeared to have upregulated levels of proteins associated with hepatocellular carcinoma. This data demonstrates the proof-of-concept of the proteomics-based biocompatibility screening approach and has identified candidate proteins and canonical pathway targets for further investigation (Figure 2). Future work will include continued refinement of the methodology and targeted validation of confirmed differentially-regulated proteins.
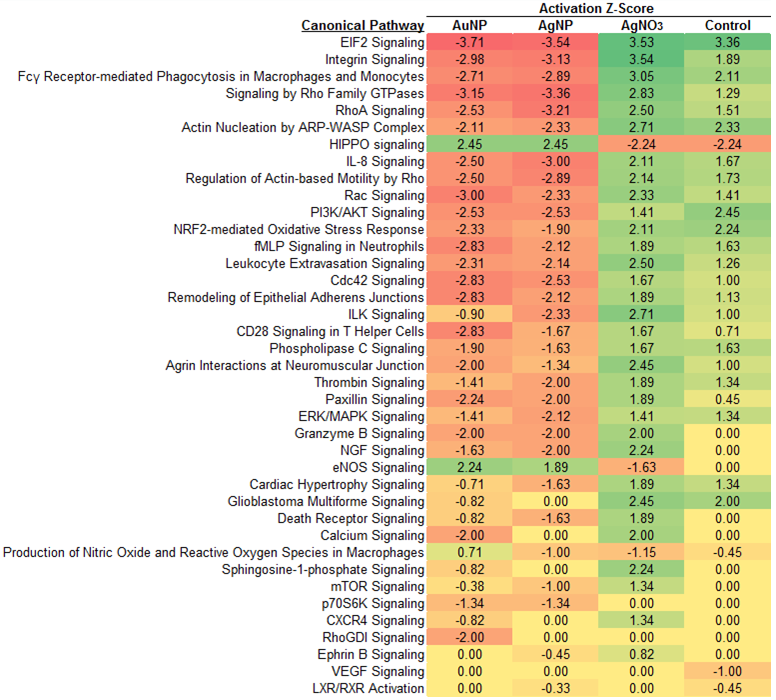
Figure 2. Results of canonical pathway analysis using Ingenuity IPA software.
Oak Ridge Institute for Science and Education
References:
[1] J.-Y. Maillard and P. Hartemann, “Silver as an antimicrobial: Facts and gaps in knowledge,” Crit. Rev. Microbiol., vol. 39, no. 4, pp. 373–383, Aug. 2012.
[2] S. W. P. Wijnhoven, W. J. G. M. Peijnenburg, C. A. Herberts, W. I. Hagens, A. G. Oomen, E. H. W. Heugens, B. Roszek, J. Bisschops, I. Gosens, D. Van De Meent, S. Dekkers, W. H. De Jong, M. van Zijverden, A. J. A. M. Sips, and R. E. Geertsma, “Nano-silver – a review of available data and knowledge gaps in human and environmental risk assessment,” Nanotoxicology, vol. 3, no. 2, pp. 109–138, Jan. 2009.
[3] A. Tebbe, M. Klammer, S. Sighart, C. Schaab, and H. Daub, “Systematic evaluation of label-free and super-SILAC quantification for proteome expression analysis,” Rapid Commun. Mass Spectrom., vol. 29, no. 9, pp. 795–801, May 2015.
Keywords:
screening,
Biocompatibility,
Nano/micro particle
Conference:
10th World Biomaterials Congress, Montréal, Canada, 17 May - 22 May, 2016.
Presentation Type:
Poster
Topic:
Safety and toxicity evaluation for biomaterials
Citation:
Narayanasamy
S,
Wickramasekara
S,
Casey
BJ and
Sussman
E
(2016). Use of discovery proteomics as a biocompatibility screening tool: study of silver nanoparticle cytotoxicity.
Front. Bioeng. Biotechnol.
Conference Abstract:
10th World Biomaterials Congress.
doi: 10.3389/conf.FBIOE.2016.01.01738
Copyright:
The abstracts in this collection have not been subject to any Frontiers peer review or checks, and are not endorsed by Frontiers.
They are made available through the Frontiers publishing platform as a service to conference organizers and presenters.
The copyright in the individual abstracts is owned by the author of each abstract or his/her employer unless otherwise stated.
Each abstract, as well as the collection of abstracts, are published under a Creative Commons CC-BY 4.0 (attribution) licence (https://creativecommons.org/licenses/by/4.0/) and may thus be reproduced, translated, adapted and be the subject of derivative works provided the authors and Frontiers are attributed.
For Frontiers’ terms and conditions please see https://www.frontiersin.org/legal/terms-and-conditions.
Received:
27 Mar 2016;
Published Online:
30 Mar 2016.
*
Correspondence:
Dr. Brendan J Casey, U.S. Food and Drug Administration, Center for Devices and Radiological Health, Silver Spring, MD, United States, brendan.casey@fda.hhs.gov